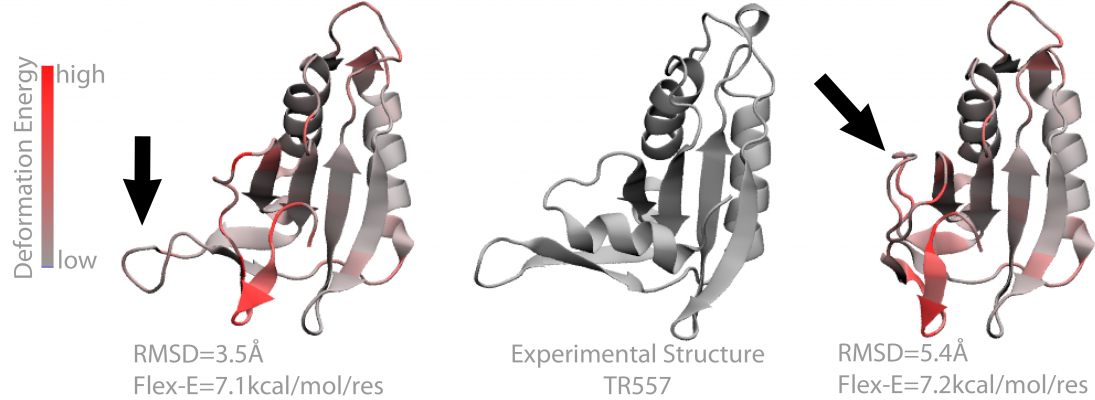
Molecular interactions dictate life. They are responsible for the structure (e.g. folding to the native state of a protein) and dynamics of macromolecules — and those in turn dictate the molecules’ functionality. Most macromolecules satisfy this functionality through interactions with other molecules. Ultimately, we would like to use this knowledge to understand the underlying causes of disease and to design new functional molecules and drugs.
We contribute to the field by developing and applying physics and AI-based tools to understand proteins, peptides, nucleic acids, and their interactions.
We are looking for graduate students and postdocs to work on these areas!
Caminante, no hay camino,
Antonio Machado
se hace camino al andar.
Traveller, there is no road,
Antonio Machado
the road is made as you walk.
Mission statement
We are here to learn and grow personally at the same time that we expand the boundaries of scientific knowledge.
We strive to produce high-impact, robust, reproducible science. The aim is that everyone who joins the group learns to: formulate hypothesis-driven research, carry it out, analyze the data, and present it in papers, and conferences, lab and department meetings. We are committed to maintaining a supportive, helpful, and respectful work environment inclusive of all, and respectful of work-life balance. We welcome new members who are aligned with fostering a healthy and supportive group dynamics and safe environment for all
News: We successfully designed new miniproteins for CAR-T cells targeting cancer as part of the BioML Challenge.
Thanks to: