Molecular Modeling and visualization have gone hand in hand over the decades. They help us understand the molecular systems we work with and correctly prepare the systems for simulation — there’s nothing worse than running a simulation for months and then finding out we missed an important protonation state or other small detail. But, don’t worry too much if it happens — we have all been there! 🙂
High School Research Experience Using VR — NSF CAREER funded
Julie and Grace joined our lab for 8 weeks through the CPET program. They learned about molecular systems we study related to cancer using virtual reality.

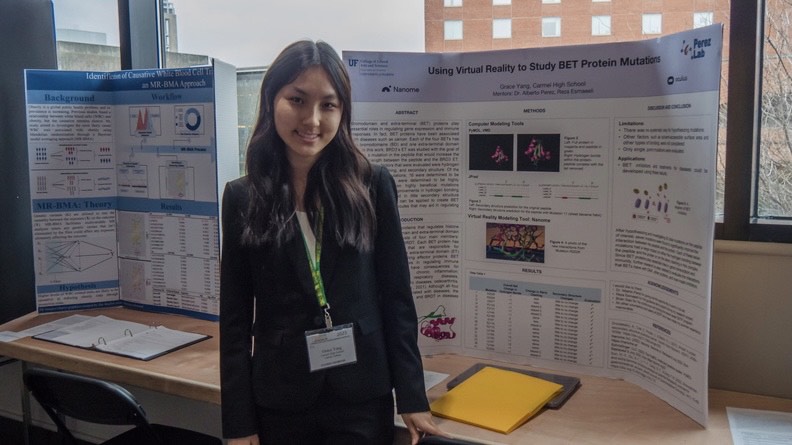
“[…]While learning about protein structure and virtual reality modeling tools, I also gained problem solving and critical thinking skills. I had an amazing time researching and using the tools at Dr. Perez’s lab. Now, eight months after the program, I have presented my summer research project at the Central Indiana Regional Science and Engineering Fair and the Hoosier Science and Engineering Fair. At those fairs, I have won the Regeneron Biomedical Science Award and Lilly’s Women in Lilly Discovery and Development Award. I have also qualified to present at the 2023 International Science and Engineering Fair in Dallas, Texas. I am so grateful to summer 2022 experience, for it has opened my eyes to aspects of science I had never encountered before in school. I have also been able to meet many incredible scientists along the way and prepare myself for my future.” Grace Yang, 2023.
Describing your system
Gareth Denyer of U of Sydney recently introduced me to sketchfab. It is a great way for sharing your system of interest in a responsive 3D visualization, with comments describing specific points you want to highlight. This is a very responsive tool for 3D in general, so it does not have the functionality of typical visualization software, but it allows users to just open a browser and follow your explanations.
Cell Paint: David Goodwill does some of the most beautiful molecular paintings I have seen, and their software allows us to participate on the process of drawing cells and viruses.
Virtual Reality
My group and I have been impressed with the immersive experience we get in a VR environment and the insights it is providing. We have been using Nanome to navigate our preferred PDB codes in ways previously unknown to us — it is very intuitive and has great functionality to rotate bonds, scale, and design/mutate proteins and nucleic acids.
We have been using Peppy developed at the University of Sidney to model peptide systems. It has a nice aspect of molecular modeling toolkits — but scales so well to the size of peptides. It even has a force field that you can run MD on while you explore the peptide’s conformations, it’s ability to adopt hairpin or helical conformations… it brings insight into molecular interactions.
Molecular visualization
VMD: good for trajectories, rendering, exporting into 3D programs and analysis — I have used this one the most.
Chimera: beautiful presets for rendering, great for static structures and cryoEM modeling.
PyMol: one of the first I worked with, some of my former group buddies loved it!
Mol*: pretty new web-based visualization. It’s worth a try!